Why bioinformatics workflows require experienced software engineers
In the world of AI, life sciences is an exciting space to work. New models and algorithms appear almost on a daily basis, helping us to understand how biology works, and how we can do things like diagnosing human health, designing new drugs, or building new microbial cell factories. It is at the core of what we do at BioLizard.
In such a fast-moving landscape, new models and algorithms appear with very different levels of maturity, from early-stage research prototypes to quickly established giants like AlphaFold.
As the field evolves, the code stacks we rely on as (bio-)informaticians evolve with it. Pipelines that once ran smoothly for years can suddenly fail because of small updates in dependent packages. Soon, you find yourself stuck in an endless cycle of updating R packages and testing each step along the way. We have all been there.
DevOps to the rescue
Before you ask, DevOps is not a housekeeping gene!
DevOps is a term used in software development and covers the combined practices of development and operations. It focuses on building, testing, and deploying software efficiently and reliably. In this context, DevOps spans several areas, including governance, automation, continuous integration and continuous delivery (CI/CD), and infrastructure as code.
Bioinformatics, however, comes with its own quirks. It is often more exploratory and experimentally driven, as researchers work toward uncovering meaningful insights while tools and analyses evolve in parallel.
In this blog post, we want to highlight what we consider most important when applying these principles to bioinformatics research and tool development.
Version 7.1_final_v3_updated
When developing analysis pipelines and experimenting with code, everything begins with proper versioning. This applies to any code-based work. Version control makes it easier to track who made changes, when they were made, and why. It also enables you to introduce new features through an appropriate branching strategy without risking the stability of your main version. And if something does break, reverting to the last stable state can be done with a single command.
The most widely used version control system today is Git¹. Git is a Distributed Version Control System² that can support projects of any size, from small scripts to large-scale software.
At BioLizard, GitHub is our preferred platform for version control, although we also work comfortably with Azure DevOps, GitLab, and others.
But it works in my environment
Creating a new pipeline that only works on your local environment will not only lead to reproducibility issues but will also make collaboration a nightmare. This is where CI/CD becomes essential.
CI/CD (Continuous Integration and Continuous Delivery/Deployment) is a set of practices designed to streamline the software development lifecycle.
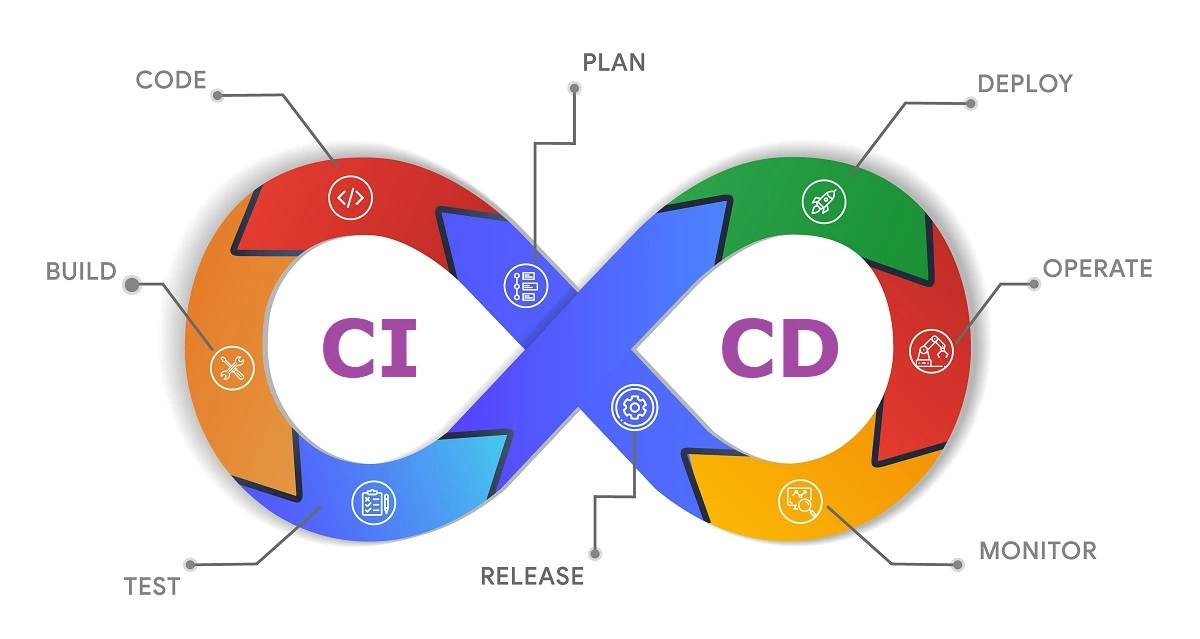
Its main purpose is to reduce the amount of manual intervention required by automating the building, testing, and deployment of software. With CI/CD pipelines in place, new code changes can be integrated and delivered quickly and reliably. This not only accelerates release cycles but also reduces the risk of human error, resulting in more stable and higher-quality applications. Ultimately, CI/CD creates a continuous flow from code commit to production, allowing teams to focus on meaningful work rather than repetitive tasks.
Continuous integration (CI) involves four steps: Plan, Code, Build and Test. Planning and Coding still rely heavily on human input (although this is changing fast with the rise of LLMs). Standardisation tools such as linters, formatters, and security checkers can support this phase. The Build and Test steps, on the other hand, can be fully automated through build systems and testing frameworks.
Continuous Development begins with Deploy, where the built and tested application is automatically placed on a server or cloud environment. The next step is Release, which makes the new version available to users, either through a manual approval (Continuous Delivery) or an immediate, automated rollout (Continuous Deployment). Once the software is live, the Operate phase ensures stability and performance. Throughout this entire process, Monitor is a crucial, ongoing activity that involves tracking key metrics and user behaviour to provide a continuous feedback loop, ensuring the system is working as intended and informing future development cycles.
At BioLizard, as a consultancy firm focused on data science and generating insights from complex datasets, our emphasis is usually on the CI part of CI/CD. To ensure the highest possible quality for our clients, we rely on a two-step code checking:
- pre-commit
- Github Actions
Pre-commit gives us fast feedback loops by checking formatting (Ruff) and security (Bandit) before each commit. This ensures that quality and security are built in from the beginning.
GitHub Actions does the same checks, but adds automated testing using Pytest. Some tests take longer to run, so we intentionally avoid including them in pre-commit in order to keep development cycles short.
We also use Vulture on an ad-hoc basis for detecting dead code, helping us keep our repositories lean and clean.
All of these tools are supported by templates, allowing our scientists to focus 100% of their time on science, uncovering new insights from complex data.
Just run it in the cloud
Using state-of-the-art tooling often requires more computational power than a local laptop can provide. For this reason, we offload large compute jobs to the cloud. There are several ways to address compute limitations, including virtual machines, batch jobs, and cloud functions. The choice depends on the specific task at hand.
Setting up a cloud environment in the UI can be tedious and is often not reproducible. Fortunately, Infrastructure as Code (IaC)³ solves this problem. IaC tools allow the entire cloud environment to be defined in configuration files that can be stored in version control. This combination is extremely powerful as it enables reproducibility, versioning of the cloud setup while also making collaboration straightforward.
Using tools such as Terraform or OpenTofu, we have created templates that allow us to spin up cloud environments quickly and securely for large compute workloads. For example, on GCP, we rely on Identity-Aware Proxy together with a Virtual Private Cloud to safeguard our virtual machines. The goal remains the same: maximise the time spent on the fun part, extracting insights from data, rather than wrestling with infrastructure setup.
This is also where the CD component of CI/CD plays a larger role. Tools like GitHub Actions enable automated deployment in addition to continuous integration. This ensures that our clients always have access to the latest version of our solutions, without manual intervention.
Conclusion
At BioLizard, science is our core. Science is only good science when it is reproducible. As we are processing large volumes of data through complex pipelines, our outcome will only be reproducible if our code is of top quality. This requires well-structured code that is consistent and maintainable.
To achieve this, we rely on a range of DevOps tools that automate critical checks and reduce the risk of human error. This automation strengthens repeatability across our workflows and frees our bioinformaticians to focus on what truly matters: generating new scientific insights rather than dealing with technical overhead.
This is just a brief look into how we approach coding and scientific work at BioLizard. Curious to learn more? Reach out, we’d be happy to explore how we can collaborate.
¹ https://rhodecode.com/blog/156/version-control-systems-popularity-in-2025
³ https://www.redhat.com/en/topics/automation/what-is-infrastructure-as-code-iac



