Breakthrough technology tackles one of science’s most complex problems
Often referred to as the molecular machinery performing the biological functions of life, the functionality of proteins largely depends on their three-dimensional shape. Because of this, accurate structure determinations are fundamental in understanding a protein’s biological functioning, its involvement in disease and its potential as a target for novel therapeutics.
However, for nearly 50 years, predicting the three-dimensional structure of proteins based on their amino acid sequence has been one of the most challenging problems in science. Typically involving complex lab analyses such as X-ray crystallography or (more recently) cryo-electron microscopy (cryo-EM), determining a single protein structure was immensely time-consuming, costly and difficult. Generations of scientists have tried to come up with computational solutions to solve the so-called “protein folding problem” for decades. They never quite succeeded, until recently – when Google’s sister company DeepMind presented AlphaFold (1).
A first, very promising version of AlphaFold was presented by the DeepMind team at the 13 biannual Critical Assessment of protein Structure Prediction (CASP13), a competition in which scientists come up with computational methods to predict (known) protein structures from their amino acid sequence.
The main breakthrough of the tool came in December of 2020, when a second and improved version of AlphaFold (AlphaFold2) was presented at CASP14. Using artificial intelligence (AI) methods, the AlphaFold2 tool was capable of producing structure predictions of a broad range of proteins with unprecedented accuracy levels. In October 2021, an addition to the tool was made that is capable of predicting the interactions within proteins composed of different subunits (so-called multimeric proteins).
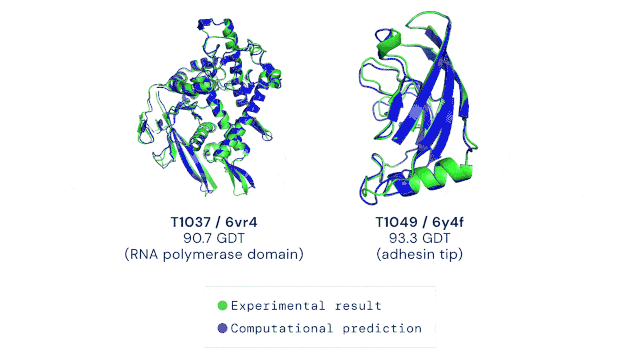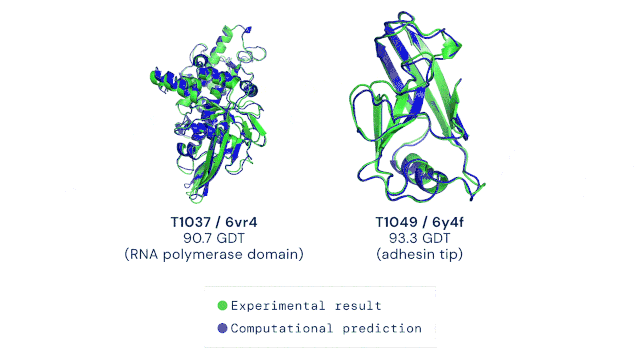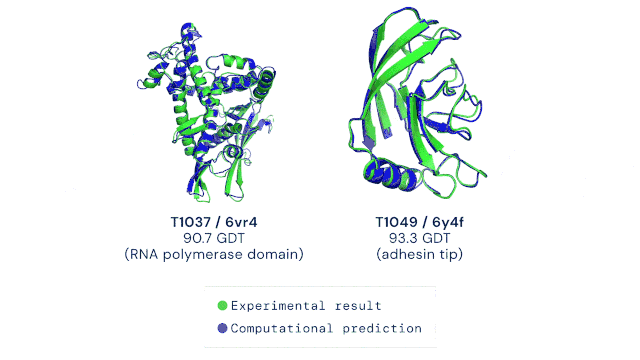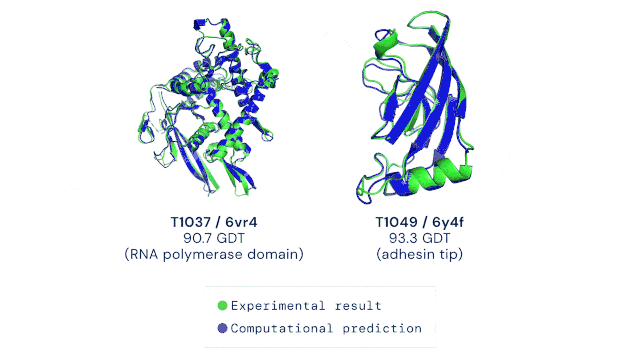
AlphaFold2 was trained on all known experimental structures stored in sequence and structure databases. The tool uses different deep learning techniques to accurately predict a protein’s structure based solely on its amino acid sequence. The ability to do this quickly and accurately, even for highly complex protein structures, is a massive leap forward in life sciences and medicine. The AlphaFold technology has, to some extent, solved the “protein solving problem” and is now pushing scientific discovery at an unprecedented pace. The DeepMind team has already predicted the structures of all available proteins from different complete genome sequences, including all known proteins in the human body (2). In 2021, Science has named AI-powered protein structure prediction as its Breakthrough of the Year (3). Moreover, the source code as well as all data for running AlphaFold2 was made freely available by the developers, allowing researchers worldwide to harness the power of AI for protein structure prediction (4). It goes without saying that the development of AlphaFold represents the dawn of an exciting new era in the life sciences.
The combination of complex computational methods to push scientific discovery has been at the centre of BioLizard’s activities from the very get-go. Our Lizards have studied AlphaFold in detail and applied the technology in a number of projects. We couldn’t be more excited about the impact this technology will have on life sciences in general and structural biology and drug development in particular.
Looking for an experienced partner in AlphaFold?
Trust the data professionals.
Read the next blog in this series here >
Sources
- Tunyasuvunakool, K., Adler, J., Wu, Z. et al. Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596 (2021). https://doi.org/10.1038/s41586-021-03828-1
- https://alphafold.ebi.ac.uk/
- https://www.science.org/content/article/breakthrough-2021
- https://github.com/deepmind/alphafold




