Microbiome analysis: Correlation is not causation
As summer turns to fall each year, we see a drop in both ice cream sales and homicides. One way to interpret this data would be to announce that you’re never eating ice cream again – it’s too dangerous! However, that interpretation has a problem: you’ve confused correlation and causation. While it’s true that ice cream sales and murder rates are positively correlated, murder isn’t caused by ice cream ingestion. These two outcomes have a common third variable, which is the real cause of both: high temperature.
As you can see, even though two points of information correlate, it does not necessarily mean that one has any influence on the other. The difference between correlation and causation is an important consideration to keep in mind when analysing the huge datasets that are involved in microbiome research.
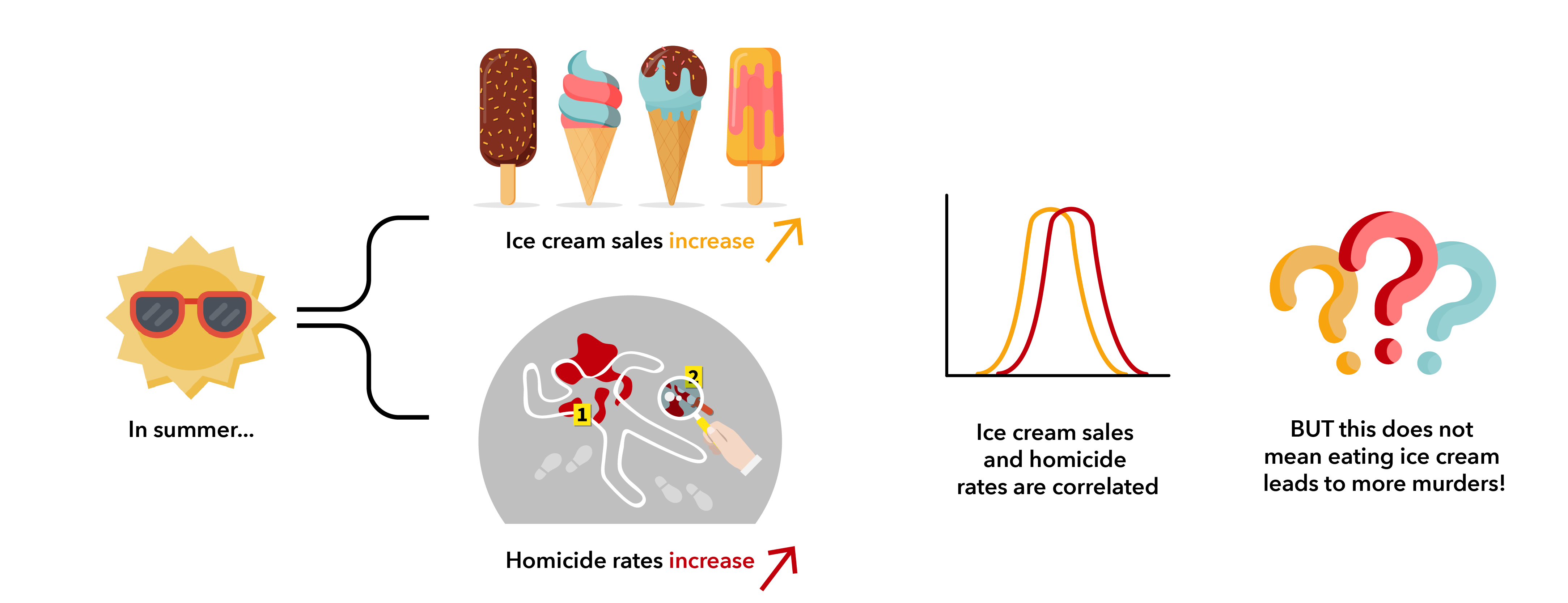
It’s pretty easy to see in the ice cream and homicide example that those two outcomes are completely unrelated events. However, it’s much harder to just use your own common sense to decipher the relationship between microbes. For instance, before announcing that you’ve found a microbe that reduces intestinal cancer rates, you’ll want to be sure that it’s not just due to pure chance, or to another unexpected third variable, that you see more of that microbe in your healthy controls versus in cancer patients. So, how and why do we approach this sticky problem of correlation versus causation in microbiome research?
Investigating causation
If causation is so difficult and dangerous to assign, why do we care about it? Well, we gave you a clue already. Let’s return to the example of a hypothetical microbe that is much more abundant in healthy controls versus in cancer patients. If it’s just a chance correlation – for instance, perhaps the microbe’s reduced abundance was due to a third variable such as the healthy controls having a different diet than the cancer patients, which exposed them to different microbes – then that microbe is not a very exciting target for further research into gut microbiology and cancer. However, if you can identify causation, for example, if you can show that patients lacking that microbe are more likely to develop cancer, you may have found an exciting new drug target. Put briefly, in biomedical fields, it’s helpful and cost-effective to get a clear picture of what’s really causing the desired outcome before you start to develop targeted drug therapies.
As you can already appreciate from the second blog in this series, there are multiple individual microbes and ecosystem processes that are at play within the complex networks of microbial communities. Furthermore, the closer the relationship becomes between two things, the harder it is to distinguish whether their relationship is correlated or causal. Correlation can be a very close type of relationship – you might find two different microbes together 99% of the time. But, that still doesn’t necessarily mean that the presence of the first microbe is causing the presence of the second. So how do we tease apart these interactions?
How to infer causation?
Today, microbiome research has largely moved beyond merely looking at what DNA is in your samples. Now, it is common practice to make use of publicly available databases to perform comparative studies. And while doing so, we can use the combined power of statistics and computational approaches to make our predictions of causation more accurate. Using such tools allows us to determine with a high accuracy, whether “this bacterium possibly causes that disease” or “if you implement this treatment, bacteria X will become more or less abundant”.
 It is possible to use statistics to test for causal relationships between whatever state you’re interested in – for example, a particular environment, disease state, or treatment – and any particular microbe or group of microbes. This will allow you to determine if it’s really causation or just a mere correlation. But we forewarned: in order to truly infer real causation, you need to be very strict about what you accept as a true causal effect between data points. In order to reduce the inevitable variability and biases in data, one good approach is to build a mechanistic model of your ecosystem.
It is possible to use statistics to test for causal relationships between whatever state you’re interested in – for example, a particular environment, disease state, or treatment – and any particular microbe or group of microbes. This will allow you to determine if it’s really causation or just a mere correlation. But we forewarned: in order to truly infer real causation, you need to be very strict about what you accept as a true causal effect between data points. In order to reduce the inevitable variability and biases in data, one good approach is to build a mechanistic model of your ecosystem.
Mechanistic models
When building a mechanistic model, or in other words an estimation of your ecosystem of interest works, there can be several factors that can explain the trends that you see. To test for true causality, we essentially run specialised statistical tests to confirm or disprove that a relationship is causal. And, to be sure of what we are dealing with, we don’t simply stop at one test. Ideally, after running 2 or 3 tests, we can arrive at a near-perfect mechanistic model of the ecosystem that will allow us to say with great certainty that, for example, intervention X is causing the disappearance of microbe Y.
Such mechanistic ecosystem models have a huge impact in pharma and biotech. For example, microbiome data from patients with Parkinson’s disease has allowed researchers to identify hundreds of bacterial species whose presence is correlated to the presence of the disease. This has provided new insights into the complex and interconnecting reasons behind Parkinson’s disease onset, and as a result, the potential for human microbiomes as both a drug target and a diagnostic tool for the disease is now being investigated. Using modern microbiome research techniques and mechanistic models for microbiome analyses meant that the scientific community was able to reach these conclusions much faster than would have been possible with older techniques. Just imagine – if laboratory researchers would have had to examine each of those hundreds of Parkinson’s disease-associated bacteria individually, for instance co-culturing them with cell cultures or organoids and waiting to see what happens, this research would have been much more time-consuming. Using a computational approach combined with advanced statistical testing allows researchers to identify the most interesting “hits” for therapeutic targeting or for use as biomarkers much more quickly.
Benefits of investigating causality
As you can guess from the examples above, there are considerable benefits to computationally investigating causality when doing microbiome research. For one, it saves a lot of time. By working with big data, you can easily tease out which bacteria are most likely to be the main causes of certain outcomes, without having to perform multiple and laborious lab tests. In addition, using computational rather than solely time- and money-intensive wet-lab approaches has also made microbiome research cheaper. As an additional bonus, once you set up an excellent analysis pipeline, you can easily apply it to future datasets, smoothing out your own future research paths. And finally, computational approaches allow you to build detailed models of ecosystems so that you can test your hypothesis in silico. Such in silico testing can also be applied to research questions that are impossible or unethical to test in a wet lab setting, giving you insights that cannot be easily obtained in any other way. Let’s stick with the example of microbiome data from a patient who has developed Parkinson’s disease, for example. We can ask, if a Parkinson’s disease patient had eaten more lactobacilli-rich foods, like yoghurt or kimchi, might they then have avoided Parkinson’s onset? This isn’t a hypothesis that we can easily or ethically test in a lab – we cannot turn back time or genetically modify humans to predispose them to Parkinson’s disease – but it is definitely a question that we can explore using mechanistic microbiome models and targeted statistical tests.
So, are you convinced that investigating causality is an exciting next step in your microbiome research line, but you’d still like some expert advice on how to implement it? Well, you’ve come to the right place. BioLizard is a true end-to-end data partner that has extensive experience in conducting microbiome research – so much so that we have even developed a new tool to help you with your microbiome data analysis process! ProBiome does the hard work on your behalf, by using state-of-the-art biostatistical frameworks combined with a user-friendly interface that generates detailed, understandable, and interactive web-based reports that allow you to gain biological insights from your microbiome datasets.








