Simulation of a mutation‘s effect on the protein structure
About the client
Our client is screening a patient‘s genome for genetic variants that are likely linked to diseases or aging. Personalised treatments are developed and tested, e.g., on cell lines derived from the patient.
About the project
Rare genetic variants may have an unknown effect, so- called “variants of unknown significance“ (VUS). The goal of this project was to study the effect of a particular VUS in a fibrous protein on the protein‘s structure and stability.
It’s a wonderful experience to work with BioLizard on bioinformatic analysis. All you need to do is to ask questions. Their strength relies on their expertise, professionalism, and their extensive network across academia and industry. No matter how difficult your questions are, they always find you the right expert who can help you solve the problem. They give me the peace of mind to stop worrying about new technology that I’m not familiar with and get back to my research goals.”
Our approach
-
Evaluating the effect of the mutation with several established effect prediction algorithms.
-
Assessing the conservation of the residue and its context in homologs in human, mammals, and other species.
-
Performing a Molecular Dynamics (MD) simulation on the wild type and the mutant structure.
Results

Position and context are highly conserved in homologs. Amino acid substitution is generally tolerated biochemically and observed in (unrelated) proteins.
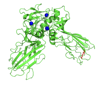
The stability of the wild type in silico mutated structure was tested in a 100 ns Molecular Dynamics simulation.
Wild type and mutant structures remain stable throughout the MD simulation. Importances is still likely due to conservation.
For more information, contact us today!